Investigating fungal diversity and uniquity
Investigating fungal diversity and uniquity based on existing and de novo data
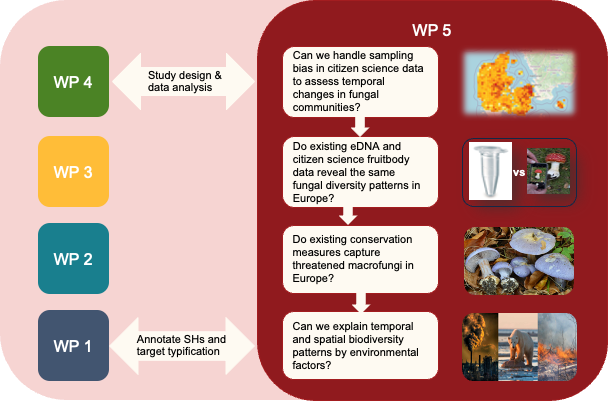
The emergence of new DNA-based techniques (WP4) and rich datasets on sporocarp records, mainly based on citizen science (WP3), now allow us to address some pressing issues on fungal biogeography and responses to global change. On one hand, citizen science data provide a higher spatiotemporal resolution than current results based on eDNA approaches, and thus have enormous potential to understand fungi under global change. On the other hand, eDNA-based approaches provide a finer and deeper taxonomic resolution (WP4). In WP5 we will explore the potentials of the two approaches for fungal monitoring and conservation, based on already existing data. We will collaborate with WP4 on several scientific objectives, and with WP1 on annotating SHs, based on the frequency and global conservation status of target species in our approach.
Objectives
- Sampling bias in citizen science data
- Same diversity patterns in Europe
- Threatened macrofungi
- Temporal and spatial biodiversity
Can sampling bias in citizen science data be handled to provide robust insight into temporal changes and spatial turnover in fungal communities? We will develop a machine learning approach to estimate sampling biases in citizen science data and implement a publicly available R or Python package handling the biases. Mapping of motivation factors in citizen science (WP3) will help us understand these biases.The resulting approach will be used for further analyses as detailed below.
Do existing eDNA and citizen science sporocarp data reveal the same fungal diversity patterns in Europe? We will explore patterns of alpha-, beta- and gamma-diversity and apply the uniquity metric to identify regions and habitat types with outstanding fungal diversity. We hypothesise that: i) eDNA and sporocarp datasets reveal the same overall diversity pattern on the European scale, as shown at national scale by ref. [10]; ii) eDNA and sporocarp datasets will show significantly different uniquity patterns due to biases specific to each approach.
Do existing conservation measures capture threatened macrofungi in Europe? We will evaluate how well the EU habitat directive and national protection schemes protect fungal uniquity and threatened fungi. We will do this both on habitat type level and by spatial overlay analysis. We hypothesise that: i) some threatened fungi are poorly captured by European conservation programmes due to associations with habitat types of limited importance to animals and plants; ii) existing eDNA data have a poor coverage of threatened fungi due to true rarity and limited spatial resolution of data.
Can we explain temporal and spatial biodiversity patterns by environmental factors? We will combine the analyses from O5.1 & 2 with spatially and temporally available environmental predictors. We will test the relation of fungal diversity metrics and changes, with environmental predictors (climate, climate change, bedrock, land use, habitat, nitrogen deposition) to explore the relative importance of the factors via effect sizes.
Tasks
Prepare data for analysis. We will download citizen data from GBIF and combine it with data obtained directly from consortium partners, including more than 1 million records from the French FongiBase not yet accessible at GBIF. We will obtain eDNA data from UNITE. Finally, environmental data (climate, soil, land use, etc.) will be achieved from Copernicus and other sources. Responsible partner: UBT in collaboration with UT, CEFE, CTFC, CSIC and UPCH.
Analyse temporal patterns. Machine learning will be applied to address sampling biases to gain robust estimates of community change over time. We will apply pertinent data cleaning procedures (e.g., coordinate cleaning) and apply down-sampling approaches of well-sampled areas to test how many records are needed to infer temporal pattern reliably. We will further compare time-series from professional well-sampled time-series (including, but not limited to Ref. [19]) with noisy citizen science time-series. Responsible partner: UBT in collaboration with CVUT, UPCH, CTFC and CEFE.
Analyse diversity patterns. Based on different datasets of eDNA and citizen science approaches (T5.1), we will analyse diversity and uniquity patterns at the European scale. Further, the biodiversity patterns will be analysed in relation to environmental factors. Using correlation analyses congruence between the approaches will be tested. Responsible partner: UBT in collaboration with CEFE, UPCH, UNIWARSAW & CTFC.
Analyse conservation status. Adding to the analyses conducted under task 5.3 we will analyse to what extent uniquity and threatened fungi are covered by European conservation initiatives, spatially as well as in relation to habitat types. Responsible partner: UPCH in collaboration with UBT, CEFE and CTFC.